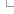Running BLAST searches
- Entering a query sequence
- Selecting the species
- Selecting the search tool
- Recent BLAST tickets
- Results
- Configuration options
BLAST is a sequence similarity search tool that can be used for both DNA and proteins. The BLAST tool compares user-supplied sequences to sequences in the WormBase ParaSite database and returns the best hits.
1) Entering a query sequence
Paste in a sequence (the suggested format is FASTA) or upload a sequence as a file. Up to 30 sequences can be added. If inputting multiple species, make sure a header (for example a FASTA header) separates each one.
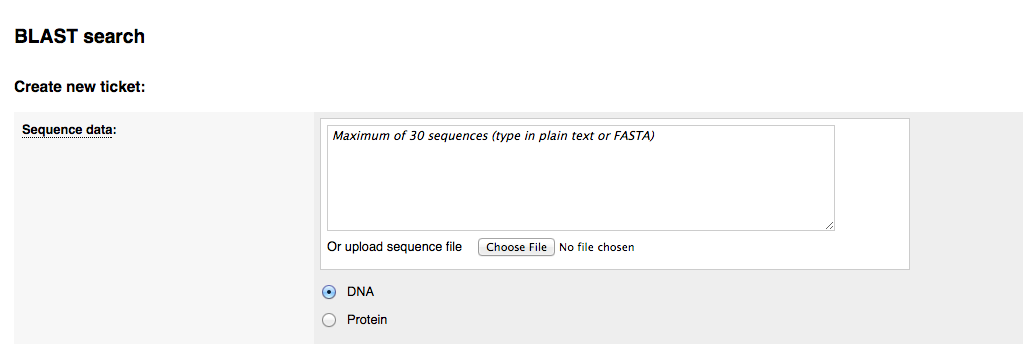
2) Selecting the species to search against
You can choose to perform the BLAST search against all genomes in ParaSite, all Nematoda, all Platyhelminthes or a custom set of species. Selecting 'Custom species list' brings up a window where you can select your species within a taxonomic tree:
DNA and protein databases available for similarity searches. When searching against the DNA database you can choose:
- Genomic sequence:
Where repetitve and/or low complexity regions are NOT masked
- Genomic sequence (hard masked):
Where genomic sequences have been run through the RepeatMasker program and repetitve and/or low complexity regions have been masked as Ns
- cDNAs (transcripts/splice variant)
3) Selecting the search tool
The following options are available, depending on the combination of query sequence and search database selected:
- BLASTN: nucleotide sequences against nucleotide databases
- BLASTX: six frame translation of a nucleotide sequence against a peptide database
- TBLASTX: translated nucleotide sequences against a translated nucleotide database
- BLASTP: peptide sequences against peptide database
- TBLASTN: peptide sequences `gainst a six frame translated nucleotide database
For BLAST searches, you can change the 'Search Sensitivity' from normal to the following:
- Near match (to find closer matches- more stringent settings than 'normal')
- Short sequences (for short sequences like primers: BLASTN only)
- Distant homologies (to allow lower-scoring pairs to pass through)
Specific parameters for these configurations can be found by expanding the Configuration options. Alternatively, change the configuration options to customise your own BLAST search.
You can give an optional description to this search in the Description field.
Once your parameters are set, click RUN to start the search.
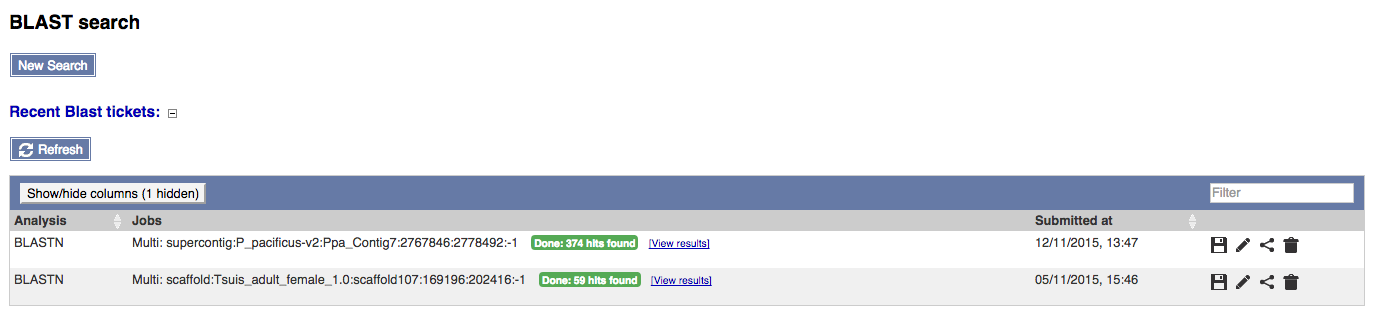
4) Recent BLAST tickets
The table lists tickets that are currently running or recently completed. You can customise the table by showing/hiding column.
The progression of the ticket gets automatically refreshed every 10 seconds until fully completed.
The icons on the right can be used to download the results of a ticket, edit and resubmit the ticket, share the ticket via URL or delete the ticket.
Click the View results link to see the results.
5) Results
The results are displayed in two sections:
- Job details:
Details of the job include job name, search type (e.g. BLASTN), date and time the job was submitted and configuration settings.
Click on the title or (-) to collaspe the Job details section.
-
Results table:
The table lists all hits in order of high to low score (and E-value) and can be customised to show/hide columns. The results can be sorted by any parameters available in the table.
Follow the links in the results table to get:
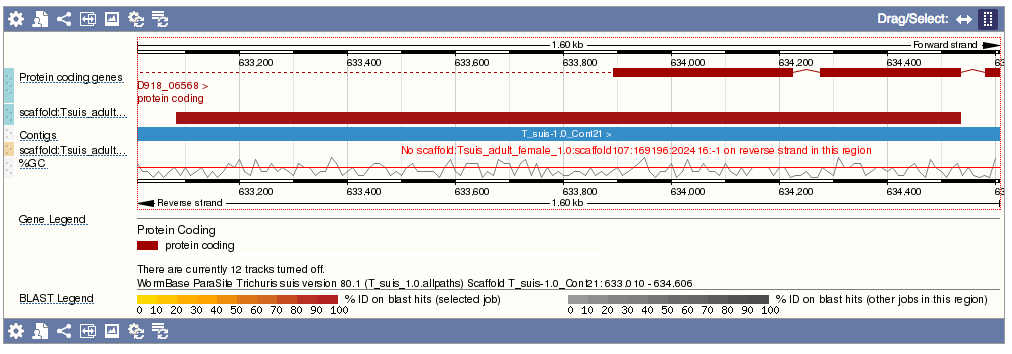
- Genomic location: shows the BLAST hit on the Region in detail view in the Location tab of the Genome Browser. The BLAST hit will appear as a red bar along the genome. You may want to click on the red bar to view a summary of the search, including E-Value, %ID, etc.
- Sequence: shows the genomic sequence or query sequence
- Alignment: shows the BLAST alignment
- Genomic location: shows the BLAST hit on the Region in detail view in the Location tab of the Genome Browser. The BLAST hit will appear as a red bar along the genome. You may want to click on the red bar to view a summary of the search, including E-Value, %ID, etc.
Click on the title or (-) to collaspe the Results table.
6) Configuration Options
General options
- Maximum number of hits to report:
- Culling limit
- E-value threshold:
Number of database hits that are displayed. The actual number of alignments may be greater than this. It varies from 10 to 5000 to 100000 and the default is 100.
This option is available for BLAST searches only, not BLAT. It varies from 1 to 999 and the default is 5. The limit will delete a hit that in enveloped by at least this many higher scoring hits.
Threshold below which results are reported. It varies from 1e-200 to 1000 and the default value is 10.