Using JBrowse displays
Table of Contents
Introduction
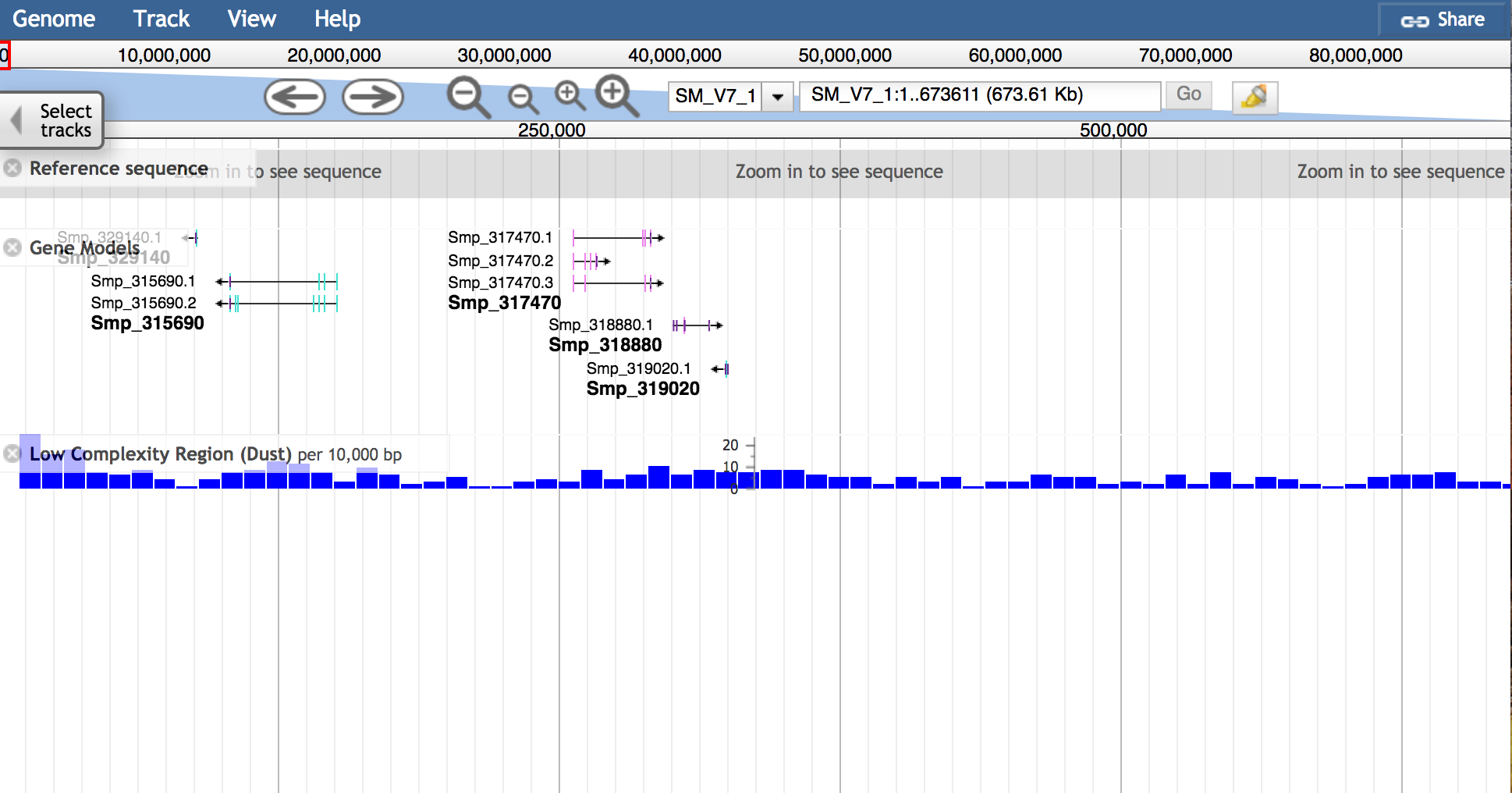
JBrowse is a fast modern genome browser that we offer as an alternative to the classic Ensembl genome browser:
Accessing JBrowse
You can navigate to the regions in JBrowse:
- from a species page, under "Navigation"
- from gene/transcript/protein etc. pages, through a "view region in JBrowse" link in the upper right corner
Basic usage
Moving
- Move the view by clicking and dragging in the track area, or by clicking left or right arrow in the navigation bar, or by pressing the left and right arrow keys.
- Center the view at a point by clicking on either the track scale bar or overview bar, or by shift-clicking in the track area.
Zooming
- Zoom in and out by clicking plus or minus symbols in the navigation bar, or by pressing the up and down arrow keys while holding down "shift".
- Select a region and zoom to it ("rubber-band" zoom) by clicking and dragging in the overview or track scale bar, or shift-clicking and dragging in the track area.
Showing Tracks
- Turn a track on by dragging its track label from the "Available Tracks" area into the genome area, or double-clicking it.
- Turn a track off by dragging its track label from the genome area back into the "Available Tracks" area.
Searching
- Jump to a feature or reference sequence by typing its name in the location box and pressing Enter.
- Jump to a specific region by typing the region into the location box as: ref:start..end.
Example Searches
- jumps to the gene WBGene0001324.
- jumps to chromosome 4
- jumps the region on chromosome 4 between 79.5Mb and 80Mb.
- centers the display at base 5,678 on the current sequence
WBGene0001324
chr4
chr4:79,500,000..80,000,000
5678
RNASeq tracks
We collaborate with the RNASeq-er project ran by the Functional Genomics Group at the European Bioinformatics Institute.
RNASeq-er retrieves public RNASeq reads of Nematoda and Platyhelminthes deposited to European Nucleotide Archive and aligns them to WormBase ParaSite genomes. We import the alignment results, retrieve additional metadata, and integrate the data into our per-species JBrowse displays.
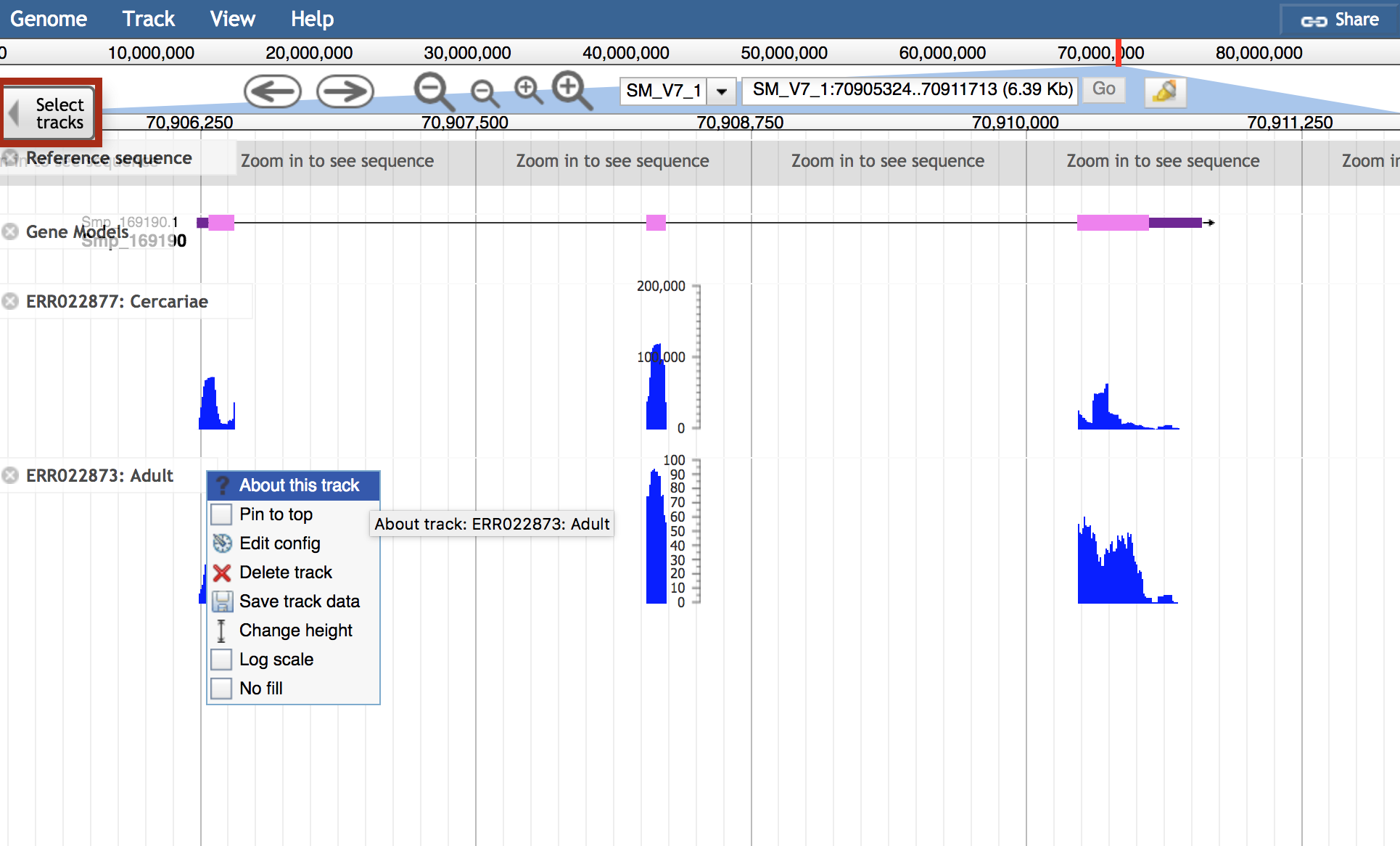
About this track in the track label menu is full of useful information about the RNASeq tracks:
- All metadata available for the track
- Links to ENA study page and other resources where available
- Link to FTP location with data files: QC, quantification, and more
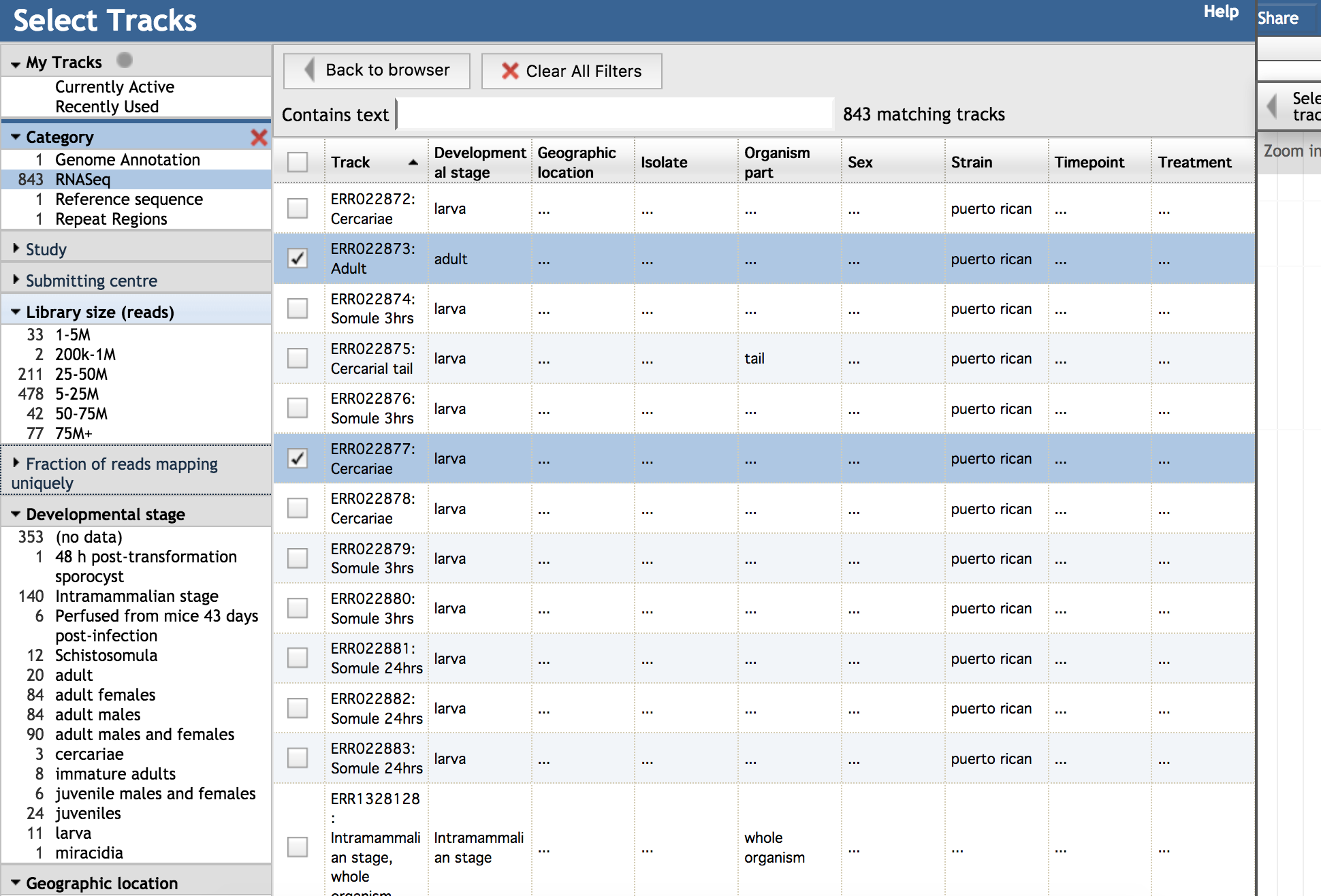
It consists of selection facets on the left, and a central grid.
The facets aggregate the runs by study, provide additional context about data quality, and organise them through several categories of metadata. The grid focuses on the metadata, and the track displays.
There are frequently too many tracks to view at once.
Select runs in the study you need, or pick several runs with good library size across several studies with good metadata.
It can also be helpful to search for keywords, and use a combination of choices across facets to narrow down your selection.